Group of Brain Development and Disease

The research group of Brain Development and Disease is driven by a fascination of the mind and its matter. The group focuses on the mechanisms involved in neurodevelopment, systemic inflammation, and neurodegenerative diseases such as dementia and Alzheimer’s disease.
We apply methods such as single cell RNA sequencing to understand the cellular diversity of the part of the brain where Alzheimer’s disease first strikes called the entorhinal cortex. Single cell RNA sequencing data has enabled us to develop a novel neuron (brain) cell type from pluripotent stem cells using an approach called direct programming. These cells may provide clues as to why some cells in the brain are more vulnerable to disease than others. Uncovering the cellular diversity of the brain provides extraordinary insight into the complexity of this organ and provides us new evidence for the function and origins of brain diseases.
Our lab is also interested in the importance of the systemic immune environment, especially the gut, its microbiota and infections, and the role these have in the onset of neurodegenerative diseases.
CURRENT PROJECTS
Global meat production continues to rise, and the human population faces future challenges in coping with rising food demand. Alternative food production chains are required to help cope with an increasing population. In-vitro grown meat also known as clean meat is an alternative food chain that serves to reduce animal production and contribute to food sustainability. However, costs in clean meat production remain high and there remains a dependency on animal biopsies for sourcing cells. This project aims to address these issues. We will use cattle embryonic stem cells to create an unlimited supply of meat cells and produce clean meat cells using cost-reducing solutions. Meat cells will be produced using a 3D cultured system incorporating edible scaffolds suitable for scaling to large volumes. Cheaper growth media will be developed by using proteins extracted from animal waste products such as manure and hair and growth factors produced from cultured cells.
Partners: DTU, Denmark and Circular Upcycling, CA, US
Funded by Independent Research Fund, Denmark.

The entorhinal cortex is a brain region that is important for processing memory and navigation. It is also the first region to be affected by Alzheimer’s disease.
In this project we are producing a cellular map of the entorhinal cortex showing neuronal spatial location, electrical activity and gene transcriptional profiles, by using state-of-the-art patch sequencing. We are also investigating sandwich lamination in the human entorhinal cortex and hope to uncover the mechanism involved.
In addition, we are studying the evolution of the spatial processing system across varying species to uncover key differences in memory and navigation at the single cell level and by using diffusion tensor imaging to demonstrate differences in connectivity of spatial processing systems to other brain regions. Together, this project aims to overturn textbook knowledge on how the brain's navigational system develops. It will provide detailed insight into the cellular map of the entorhinal cortex and how navigation and memory processing differs between species.

Funded by Lundbeck Foundation
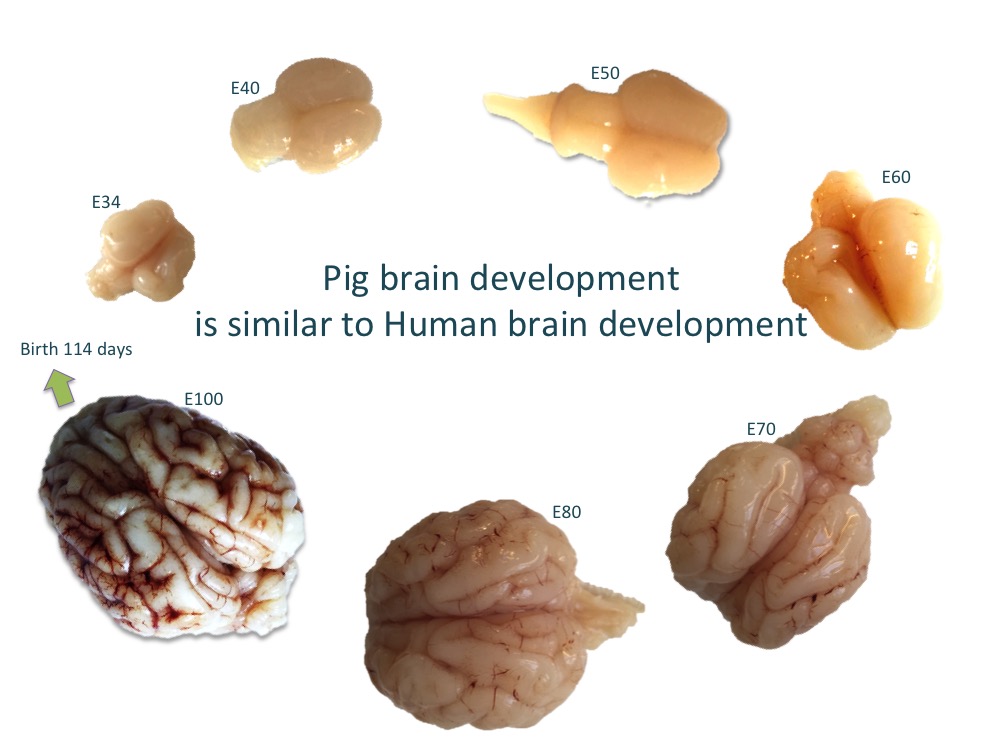
We are particularly interested in the role of the entorhinal cortex in Alzheimer’s disease and why it is one of the first regions of the brain that is affected by the disease. In this project, we have performed detailed investigation of the anatomical and molecular make-up of the entorhinal cortex using the developing pig brain and are using state-of-the-art single-cell RNA sequencing technology to discover more details on the stellate cell located in the entorhinal cortex and its role in the disease. The aim is to produce a stellate cell in the dish from human induced pluripotent stem cells and study why it is particularly susceptible in Alzheimer’s disease.

 Funded by Innovation Foundation (Brainstem) and The Danish Council for Independent Research: Technology and Production
Funded by Innovation Foundation (Brainstem) and The Danish Council for Independent Research: Technology and Production
The cell is the fundamental unit in biology and thanks to technological advances, it is now possible to profile individual cells. However, to understand the function of complex tissues both in health and disease, it is necessary to (i) profile several aspects of each cell, (ii) understand its location, and (iii) understand functional cell-cell interactions.
In this project, we are developing a novel technology, SPAtIal CONnectomics (SpaiCon), which can process complex cellular information. We are doing so by combining the multiplexed error-robust fluorescence in situ hybridization (MERFISH) platform, for spatial transcriptomics profiling with multielectrode arrays for measuring electrophysiological activity. MERFISH characterizes gene expression in individual cells based on fluorescent probes and records the spatial location of transcripts from cell/tissue slices. Multielectrode arrays record and/or stimulate the electrical activity of cells at specific sites within a piece of tissue, making it possible to infer spatiotemporal dynamics. We are merging our diverse expertise from the fields of microelectronics, neuroscience and bioinformatics to create a novel platform that merges these technologies into a novel multi-omic technology. This project involves manufacturing and testing of custom built transparent multielectrode arrays that are compatible with MERFISH technology, thus generating a new computational workflow to merge and analyse the complex data.
SpaiCon will be transformative not only to neuroscience, but also to cardiology and other areas of biology as it will provide an unprecedented characterization of tissues. In particular, it will open up new possibilities for understanding how gene expression influences electrophysiological activity, and vice versa. We anticipate that SpaiCon will be of interest to researchers studying basic biology, tissue regeneration and vascularization and to translational researchers developing novel therapies for varying diseases.
Partners: Brigham and Womens Hospital, Boston, US and Universite de Lille, France
Funded by Novo Nordisk Foundation

Recent research outputs
This community based paper published in Nature Neuroscience voices the need to catalogue cellular diversity in the brain using a unified taxonomy including single cell transcriptomics and other data obtained from different approaches. https://www.nature.com/articles/s41593-020-0685-8
This review highlights the progress that pluripotent stem cells have made in being able to remodel complex tissue types such as the blood-brain barrier.
Paving the way towards complex blood-brain barrier models using pluripotent stem cells.
This study was the first to produce an in-vitro pig cell model of Alzheimer’s disease. Neural progenitors were studied for early phenotype of the disease and several mechanisms related to AD pathology could be seen.
Embryonic stem cell-derived radial glial cells from the APPsw transgenic pig show altered APP activity and tau splicing as well as perturbations in the cell cycle.
It is challenging to produce bona-fide iPSCs in the pig. In this study, we found a subpopulation of cells in pig fibroblasts that are reprogrammable, whilst other fibroblasts are not, suggesting that reprogramming success is critically dependent on the characteristics of the original cells.
Identification of SSEA-1 expressing enhanced reprogramming (SEER) cells in porcine embryonic fibroblasts.
We recently have created a novel pluripotent stem cell-derived model of the entorhinal cortex. Using a forward programming approach, we were able to overexpress 6 transcription factors in human induced pluripotent stem cells to produce stellate-cell like cells expressing Reelin. We anticipate this cellular model will be of use for studying early mechanisms in disease such as Alzheimer’s disease. We further characterize the developing and postnatal porcine medial entorhinal cortex using single cell RNA sequencing. In press at Frontiers in Cell and Developmental Biology
A recent article from the group highlights the anatomical characterization of the developing and adult porcine entorhinal cortex. Using BrdU labeling in the developing mouse entorhinal cortex we identify that the lamination of the entorhinal cortex occurs by parallel development of the deep layers followed by parallel development of the superficial layers. Development of the Entorhinal Cortex Occurs via Parallel Lamination During Neurogenesis
- Hawrylycz, M., Aalling, N., Arendt, D., Armananzas, R., Ascoli, G. , Bielza, C., Bokharaie, V., Bergmann, T., Bystron, I., Capogna, M., Chang, Y., Clemens, A., de Kock, C., DeFelipe, J., Dos santos, S.E , Dunville, K., Feldmeyer, D., Fiáth, R., Fishell, G., Foggetti, A., Gao, X., Ghaderi, P., Güntürkün, O., Hall, V.J., Helmstaedter, M., Herculano, S., Hilscher, Hirase, H., Hjerling-Leffler, J., Hodge, R., Huang, J., Huda, R., Juan, Y., Khodosevich, K., Kiehn, O., Koch, H., Kuebler, E., Kühnemund, M., Larrañaga, P., Lelieveldt, B., Louth, E.L., Lui, J., Mansvelder, H., Marin, O., Martínez-Trujillo, J., Moradi, H., Goriounova, N., Mohapatra, A., Nedergaard, M., Němec, P., Ofer, N., Pfisterer, U., Pontes, S., Redmond, W., Rossier, J., Sanes, J., Scheuermann, R., Serrano Saiz , E., Somogyi , P., Tamás, G., Tolias, A., Tosches, M., Turrero Garcia, M., Aguilar-Valles, A., Munguba, H., Wozny, C., Wuttke, T., Yong, L. , Zeng, H., Lein, E.S. (2020) A community-based transcriptomics classification and nomenclature of neocortical cell types. Nature Neuroscience 2020 IF. 20.071. doi: 10.1038/s41593-020-0685-8.
- Nyhus, C., Pihl, M., Hyttel, P., Hall, V.J. (2019) Evidence for nucleolar dysfunction in Alzheimer’s disease. Reviews in the Neurosciences: 30:7 IF 3.078.
- Nawzad Aubid, N., Liu, Y., Peralvo Vidal, J.M., and Hall, V.J. (2019) Isolation and culture of porcine primary fetal progenitors and neurons from the developing dorsal telencephalon. Journal of Veterinary Science. Mar;20(2):e3
- Li, D., Secher, J., Hyttel, P., Ivask, M., Kolko, M., Hall, V.J*., Freude, K.K*. (2018) Generation of transgene-free porcine intermediate type induced pluripotent stem cells. Cell Cycle. Accepted. IF 3.952 *equal senior author
- Bernardo, A.S., Jouneau, A., Marks, H., Kensche, P., Kobolak, J., Freude, K., Hall, V., Feher, A., Polgar, Z., Sartori, C., Bock, I., Louet, C., Faial, T., Kerstens, H.H.D., Bouissou, C., Parsonage, G., Mashayekhi, K., Smith, J.C., Lazzari, G., Hyttel, P., Stunnenberg, H.G., Huynen, M., Pedersen, R.A., Dinnyes, A. (2018) Mammalian embryo comparison identifies novel pluripotency genes associated with the naive or primed state. Biol Open. 17;7(8)
- Chandrasekaran, A., Avci, HX, Ochalek, A, Rosingh, LN., Molnar, K., Laszlo, L., Bellak, T., Teglasi, A., Pesti, K., Mike A., Phanthong, P, Biro, O., Hall, V., Kitiyanant, N., Krause, K.H., Kobolak, J., Dinneyes, A. (2017). Comparison of 2D and 3D neural induction methods for the generation of neural progenitor cells from human induced pluripotent stem cells. Stem Cell Res. 25:139-151.
- Lauschke, K., Frederiksen, L., and Hall, V.J. (2017). Paving the way towards complex blood-brain barrier models using pluripotent stem cells. Stem cells and Development. 26(12):857-874.
- Li, D., Secher, J.O., Juhl, M., Mashayekhi, K., Nielsen, T.T., Holst, B., Hyttel, P., Freude, K.K. and Hall, V.J. (2017) Identification of SSEA-1 expressing enhanced reprogramming (SEER) cells in porcine embryonic fibroblasts. Cell Cycle. 16(11):1070-1084.
- Tubsuwan, A., Pires, C., Rasmussen, M.A., Schmid, B., Nielsen, J.E., Hjermind, L.E., Hall, V., Nielsen, T.T., Waldemar, G., Hyttel, P. et al (2016) Generation of induced pluripotent stem cells (iPSCs) from an Alzheimer’s disease patient carrying a L150P mutation in PSEN-1. Stem cell research. 16(1): 110-112.
- Hall, V.J., Lindblad, M.M., Jakobsen, J.E., Gunnarsson, A., Schmidt, M., Rasmussen, M.A., Volke, D., Zuchner, T., Hyttel, P. (2015) Embryonic stem cell-derived radial glial cells from the APPsw transgenic pig show altered APP activity and tau splicing as well as perturbations in the cell cycle. Disease Models and Mechanisms 8(10):1265-78.
- Freude, K., Pires, C., Hyttel, P. and Hall, V.J. (2014) Induced pluripotent stem cells derived from Alzheimer’s disease patients: the promise, the hope and the path ahead. Journal of Clinical Medicine. 3(4):1402-36.
- Hall, V.J. and Hyttel, P. (2014) Breaking down pluripotency in the porcine embryo reveals both a premature and reticent stem cell state in the inner cell mass and unique expression profiles of the naïve and primed stem cell states. Stem Cells and Development. 23(17):2030-45.
- Hall, V.J., Kristensen, M., Rasmussen, M.A., Ujhelly, O., Dinnyes, A., Hyttel, P. (2012). Temporal repression of endogenous pluripotency genes during reprogramming of porcine induced pluripotent stem cells. Cellular Reprogramming. 14(3):204-216.
- Yu, G., Jammes, H., Rasmussen, MA., Oestrup, O., Beaujean, N., Hall, VJ., Hyttel, P. (2011) Epigenetic regulation of gene expression in porcine epiblast, hypoblast and trophectoderm and epiblast derived neural progenitor cells. Epigenetics. 6(9):1149-1161.
- Rasmussen, M.A., Hall, V.J., Carter, T.F., Hyttel, P. (2011). Directed differentiation of porcine epiblast-derived neural progenitor cells into neurons and glia. Stem Cell Research 7(2):124-136.
- Yu, G., Hyttel, P, and Hall, V.J. (2011). Dynamic changes in epigenetic marks and gene expression during porcine epiblast specification. Cellular Reprogramming. 13(4):345-360.
Group leader
Associate Professor Vanessa Hall
Grønnegårdsvej 7,
DK-1870 Frederiksberg
Tel: +45 35332512
E-mail: vh@sund.ku.dk
GROUP MEMBERS
Vanessa Hall, Associate professor vh@sund.ku.dk,, Group leader
Dorottya Ralbovszki, PhD student, dmr@sund.ku.dk
Selma Olsen, master’s student, tausen@ruc.dk
Shanna Ninett Christensen, master’s student, fwn895@alumni.ku.dk
Kirtika Kumar, master’s student, hsw781@alumni.ku.dk
Eliska Waloschková, Postdoctoral researcher, ew@sund.ku.dk
Jesse Westfall, PhD student jjwe@sund.ku.dk
EMERITUS GROUP MEMBER
Tobias Bergmann, (emeritus PhD student)
Yong Liu, (emeritus PhD student)
